Exploring the Power of Transformers in 3D Medical Image Segmentation: A Deep Dive into UNETR
In recent years, transformers have emerged as a groundbreaking trend in the field of computer vision, particularly in medical imaging. Their ability to capture long-range dependencies and contextual information has made them a compelling alternative to traditional convolutional neural networks (CNNs). In this article, I will delve into my re-implementation of a transformer-based model for 3D segmentation, specifically focusing on the UNETR architecture. We will compare its performance against the classical UNET model using the BRATS dataset, which contains 3D MRI brain images.
Understanding UNETR: A Transformer for 3D Medical Image Segmentation
UNETR, introduced by Hatamizadeh et al., is the first successful transformer architecture designed explicitly for 3D medical image segmentation. This model leverages the strengths of transformers to enhance the segmentation capabilities of traditional architectures like UNET. The BRATS dataset, which we will use for our experiments, is a multi-modal large-scale 3D imaging dataset that provides a rich source of MRI images captured under various modalities.
Overview of the BRATS Dataset
The BRATS dataset is particularly challenging due to its focus on tumor segmentation. It contains four 3D volumes of MRI images, with annotations limited to the tumor regions. This makes the segmentation task more complex, as the model must accurately localize the tumor amidst varying intensities and structures in the images. The dataset categorizes tumors into three main types:
- Edema: The whole tumor, typically visible in T2-FLAIR MRI images.
- Non-enhancing solid core: The tumor core, visible in T2 MRI.
- Enhancing tumor structures: Usually visible in T1Gd, surrounding the necrotic core.
Data Loading and Transformation with MONAI
To facilitate our experiments, I utilized the MONAI library, an open-source framework designed for medical imaging. MONAI simplifies the process of loading datasets and applying necessary transformations. Using the DecathlonDataset class, we can easily load the BRATS dataset and apply a series of transformations to prepare the data for training.
from monai.apps import DecathlonDataset
train_ds = DecathlonDataset(
root_dir=root_dir,
task="Task01_BrainTumour",
transform=train_transform,
section="training",
download=True,
num_workers=4,
cache_num=8,
)The transformation pipeline is crucial for ensuring that the model receives data in a format that enhances its learning capabilities. We resample the images to a voxel size of 1.5, 1.5, and 2.0 mm in each dimension and apply random 3D sub-volumes of sizes 128, 128, and 64. Additionally, we implement augmentations such as random flipping and intensity jittering to improve the model’s robustness.
The UNETR Architecture
The UNETR architecture integrates transformers into the traditional UNET framework, allowing for improved feature extraction and segmentation accuracy. The architecture consists of an encoder-decoder structure, where the encoder utilizes transformers to capture global context, and the decoder reconstructs the segmentation map.
from self_attention_cv import UNETR
model = UNETR(
img_shape=tuple(roi_size),
input_dim=4,
output_dim=3,
embed_dim=512,
patch_size=16,
num_heads=10,
ext_layers=[3, 6, 9, 12],
norm='instance',
base_filters=16,
dim_linear_block=2048
).to(device)This model boasts approximately 49.7 million parameters, making it a powerful contender in the realm of 3D medical image segmentation.
Training the Model
For training, we employed the DICE loss combined with cross-entropy, which is particularly effective for segmentation tasks. The training loop iterates through the dataset, optimizing the model parameters based on the computed loss.
loss_function = DiceCELoss(to_onehot_y=False, sigmoid=True)
optimizer = torch.optim.AdamW(model.parameters(), lr=1e-4, weight_decay=1e-5)
for epoch in range(max_epochs):
model.train()
for batch_data in train_loader:
inputs, labels = (batch_data["image"].to(device), batch_data["label"].to(device))
optimizer.zero_grad()
outputs = model(inputs)
loss = loss_function(outputs, labels)
loss.backward()
optimizer.step()Baseline Comparison: UNET
To evaluate the performance of UNETR, we compared it against a well-configured UNET model. The results were promising, with UNETR achieving a mean DICE coefficient of 76.9%, slightly outperforming the UNET baseline at 76.6%. This comparison highlights the potential of transformer architectures in enhancing segmentation tasks.
| Model | Epochs | Mean DICE Coefficient |
|---|---|---|
| UNET (baseline) | 170 | 76.6% |
| UNETR (self-attention-cv) | 180 | 76.9% |
| UNETR (MONAI) | 180 | 76.1% |
Visualizing the Results
The effectiveness of the models can be further illustrated by comparing the output segmentation maps against the ground truth annotations. The visualizations reveal that UNETR produces smoother and more accurate segmentations, particularly in complex regions where tumors overlap.
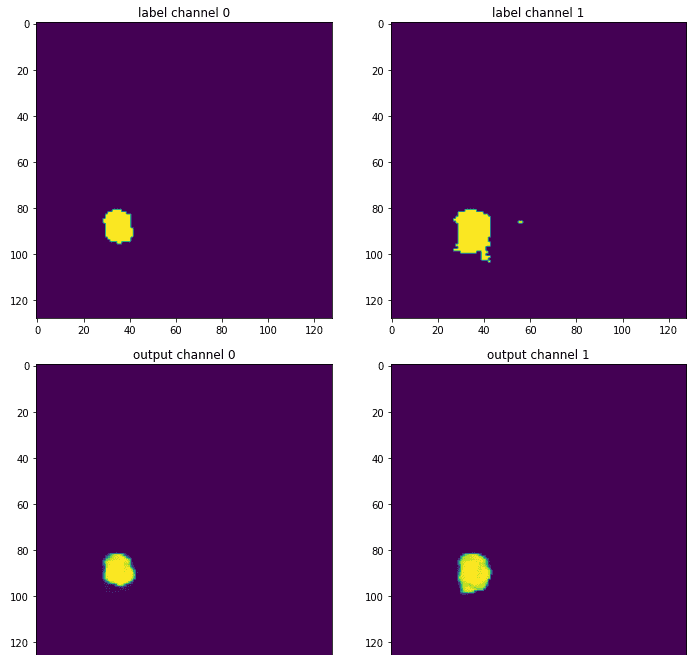
Conclusion and Future Directions
While the results from the UNETR model are encouraging, I remain cautious about the overall performance of transformers in 3D medical imaging. The success of these models often hinges on the quality of data preprocessing and transformation pipelines, which can overshadow the architectural innovations. As the field continues to evolve, I anticipate further advancements that will refine these models and enhance their applicability in medical imaging.
In summary, the exploration of transformer architectures like UNETR presents exciting opportunities for improving segmentation tasks in medical imaging. As researchers continue to innovate and optimize these models, we may witness a paradigm shift in how we approach complex imaging challenges.
For those interested in furthering their understanding of deep learning in production, I invite you to check out our book on Deep Learning in Production, which provides insights into deploying and scaling machine learning models effectively.
Thank you for your interest in AI, and stay tuned for more updates and insights into the world of medical imaging and deep learning!
