Exploring U-Shaped Architectures: The Evolution of U-Net in Medical Image Segmentation
In the realm of deep learning, particularly in the field of computer vision, U-shaped architectures have emerged as a powerful tool for tasks such as image segmentation. These architectures, characterized by their encoder-decoder structure, have proven to be exceptionally effective in various real-world applications, especially in the medical domain. This article delves into the intricacies of U-Net architectures, tracing their evolution and highlighting their significance in medical image analysis.
Understanding U-Shaped Architectures
At the core of U-shaped architectures lies a specific encoder-decoder scheme. The encoder progressively reduces the spatial dimensions of the input while increasing the number of channels, effectively capturing high-level features. Conversely, the decoder increases the spatial dimensions while reducing the number of channels, ultimately restoring the original input size to make pixel-wise predictions. The tensor that passes through the decoder is often referred to as the "bottleneck," which contains the most compressed representation of the input data.
This unique structure allows U-shaped models to excel in tasks requiring precise localization, such as semantic segmentation, where the goal is to classify each pixel in an image. The U-Net architecture, in particular, has stood the test of time and remains a benchmark for many segmentation tasks.
The Fully Convolutional Network (FCN)
The journey of U-shaped architectures began with the Fully Convolutional Network (FCN), one of the first architectures to eliminate fully connected layers. FCNs can be trained end-to-end and are capable of processing inputs of arbitrary sizes, making them versatile for pixel-level predictions. The architecture employs transposed convolutions as a trainable upsampling method, allowing it to generate high-resolution outputs from low-resolution inputs.
FCN Implementation Example
Here’s a simplified implementation of an FCN using PyTorch:
import torch
import torch.nn as nn
class FCN32s(nn.Module):
def __init__(self, pretrained_net, n_class):
super().__init__()
self.n_class = n_class
self.pretrained_net = pretrained_net
self.relu = nn.ReLU(inplace=True)
self.deconv1 = nn.ConvTranspose2d(512, 512, kernel_size=3, stride=2, padding=1, output_padding=1)
self.bn1 = nn.BatchNorm2d(512)
# Additional deconvolutional layers...
self.classifier = nn.Conv2d(32, n_class, kernel_size=1)
def forward(self, x):
output = self.pretrained_net(x)
x5 = output['x5']
score = self.bn1(self.relu(self.deconv1(x5)))
# Additional forward passes...
score = self.classifier(score)
return scoreU-Net and 3D U-Net
Building upon the foundation laid by FCNs, the U-Net architecture introduced the concept of skip connections, which allow high-resolution features from the encoder to be combined with the upsampled outputs in the decoder. This design choice enhances the model’s ability to localize segmentations accurately.
U-Net Architecture Overview
The U-Net architecture can be divided into two main paths: the encoder (contracting path) and the decoder (expansive path). The encoder consists of repeated applications of convolutional layers followed by max pooling, while the decoder employs upsampling and concatenation with corresponding encoder features.
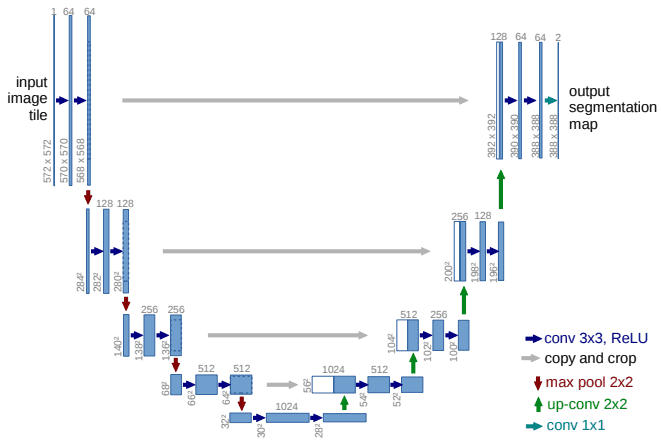
Implementation of 2D U-Net
Here’s a basic implementation of the U-Net architecture:
import torch
import torch.nn as nn
import torch.nn.functional as F
class DoubleConv(nn.Module):
def __init__(self, in_ch, out_ch):
super(DoubleConv, self).__init__()
self.conv = nn.Sequential(
nn.Conv2d(in_ch, out_ch, 3, padding=1),
nn.BatchNorm2d(out_ch),
nn.ReLU(inplace=True),
nn.Conv2d(out_ch, out_ch, 3, padding=1),
nn.BatchNorm2d(out_ch),
nn.ReLU(inplace=True)
)
def forward(self, x):
return self.conv(x)
# Additional classes for InConv, Down, Up, and OutConv...
class Unet(nn.Module):
def __init__(self, in_channels, classes):
super(Unet, self).__init__()
self.inc = InConv(in_channels, 64)
self.down1 = Down(64, 128)
# Additional layers...
self.outc = OutConv(64, classes)
def forward(self, x):
x1 = self.inc(x)
x2 = self.down1(x1)
# Additional forward passes...
x = self.outc(x)
return xThe 3D U-Net
The 3D U-Net was introduced to handle volumetric data, such as MRI scans. It extends the U-Net architecture by applying 3D convolutions, allowing it to capture spatial relationships in three dimensions effectively.
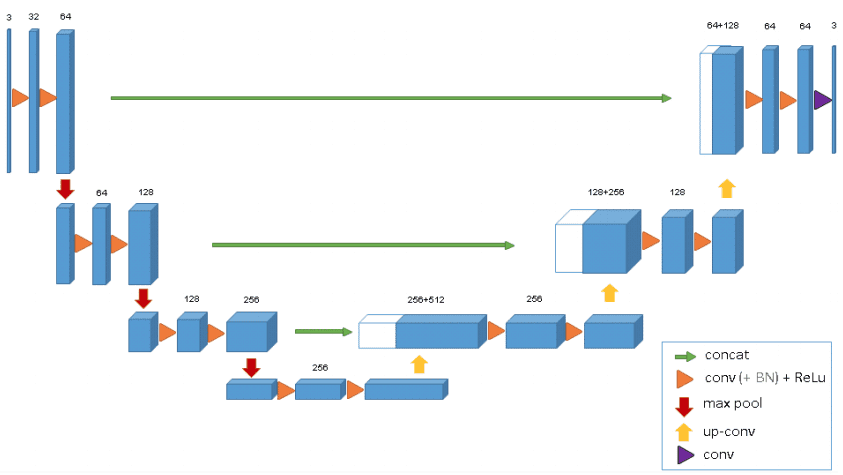
Advanced Variants of U-Net
As the field of medical image segmentation has evolved, several advanced variants of U-Net have been developed to enhance performance and address specific challenges:
V-Net (2016)
V-Net extends U-Net for 3D MRI volume segmentation, utilizing 3D convolutions instead of processing slices individually. It employs strided convolutions for downsampling and introduces residual connections to improve gradient flow.
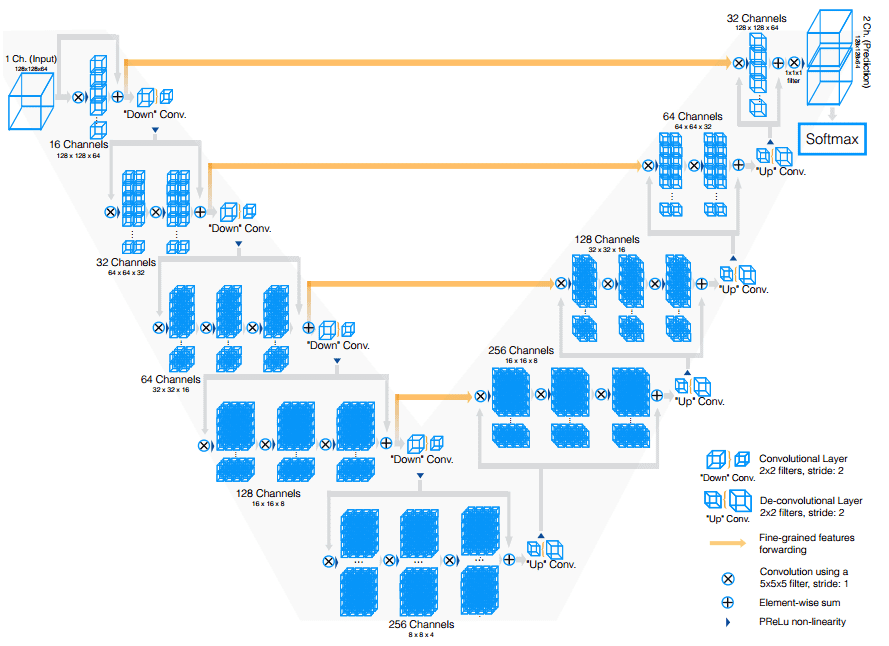
UNet++ (2018)
UNet++ introduces nested skip pathways to bridge the semantic gap between encoder and decoder feature maps. This architecture enhances the model’s ability to capture fine-grained details.
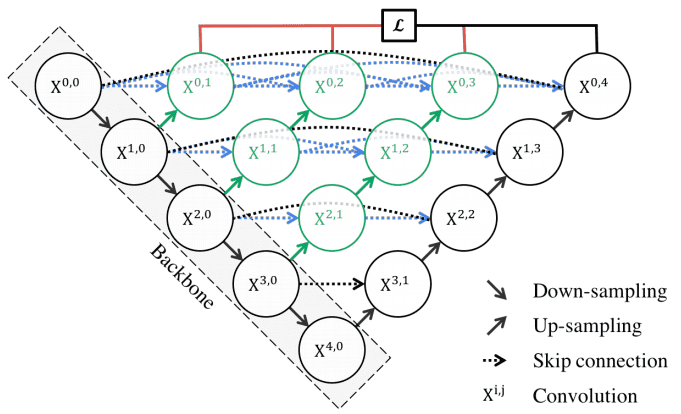
MultiResUNet (2020)
MultiResUNet incorporates inception-like modules to capture features at multiple resolutions, addressing the challenges posed by multimodal medical images.
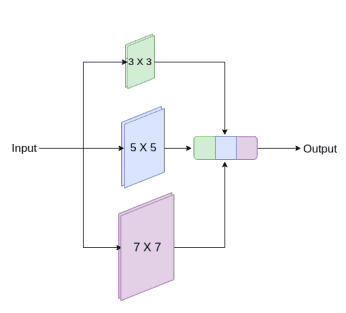
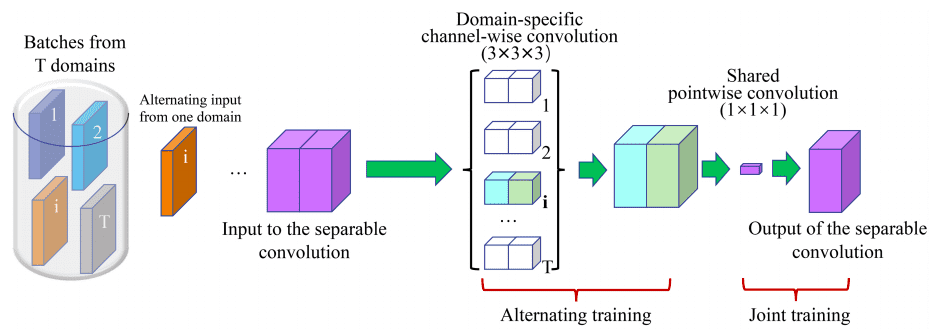
3D U²-Net
The 3D U²-Net employs channel-wise separable convolutions to reduce the number of parameters while maintaining performance across various domains.
Conclusion
The evolution of U-shaped architectures, particularly U-Net and its variants, has significantly impacted the field of medical image segmentation. These models have demonstrated remarkable success in accurately segmenting complex structures in medical images, paving the way for advancements in diagnostics and treatment planning.
As the field continues to evolve, researchers and practitioners are encouraged to explore these architectures and their adaptations to tackle specific challenges in medical imaging. For those interested in delving deeper into the intersection of AI and medicine, the AI for Medicine course on Coursera offers valuable insights and knowledge.
References
- Long, J., Shelhamer, E., & Darrell, T. (2015). Fully convolutional networks for semantic segmentation.
- Ronneberger, O., Fischer, P., & Brox, T. (2015). U-net: Convolutional networks for biomedical image segmentation.
- Milletari, F., Navab, N., & Ahmadi, S. A. (2016). V-net: Fully convolutional neural networks for volumetric medical image segmentation.
- Çiçek, Ö., Abdulkadir, A., Lienkamp, S. S., Brox, T., & Ronneberger, O. (2016). 3D U-Net: learning dense volumetric segmentation from sparse annotation.
- Zhou, Z., Siddiquee, M. M. R., Tajbakhsh, N., & Liang, J. (2018). Unet++: A nested u-net architecture for medical image segmentation.
- Ibtehaz, N., & Rahman, M. S. (2020). MultiResUNet: Rethinking the U-Net architecture for multimodal biomedical image segmentation.
- Huang, C., Han, H., Yao, Q., Zhu, S., & Zhou, S. K. (2019). 3D U²-Net: A 3D Universal U-Net for Multi-domain Medical Image Segmentation.
By understanding and leveraging these architectures, we can continue to push the boundaries of what is possible in medical image analysis, ultimately improving patient outcomes and advancing the field of medicine.
